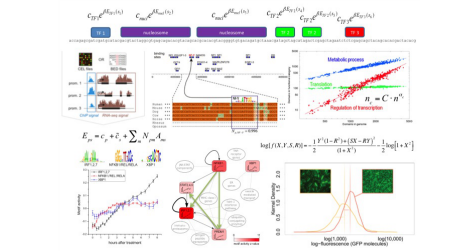
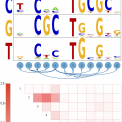
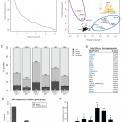
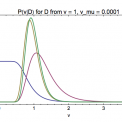
Welcome to Erik van Nimwegen's lab page at the Biozentrum of the University of Basel. We are a group of interdisciplinary researchers with backgrounds ranging from theoretical physics to molecular biology that are studying the function and evolution of the regulatory networks that cells use to control the expression of their genes. Typical questions that we are trying to address include: How are the regulatory networks encoded into the genome sequence? How is this regulatory code read out by the cell? How have these regulatory networks evolved? And, are there general `design principles' to these regulatory networks? Another key interest of our group is the identification of general quantitative laws in genome evolution.

Research
Most projects that we pursue concern the functioning and evolution of genome-wide regulatory systems in organisms ranging from bacteria to humans.
Resources
Our lab has developed a range of software tools, web-services, and databases in regulatory and comparative genomics. Besides links to these tools the resources page also provides links to teaching materials and presentations from our group.

Group
We are an interdisciplinary group of researchers with backgrounds ranging from theoretical physics and computer science to molecular biology.

Contact & Positions
Biozentrum, University of BaselKLB61, Room 916
Phone: +41 61 267 15 86
Fax: +41 61 267 15 85
Email: yvonne.steger-at-unibas.ch