Main Content
Uropathogenic Escherichia coli (UPEC) and urinary tract infections (UTIs)
Deciphering UPEC in-vivo physiology
To truly understand how UPEC behave inside the human bladder, we go straight to the source – analyzing urine samples from UTI patients at University Hospital Basel (in collaboration with Sarah Tschudin).
Our multi-pronged approach includes:
- In-vivo proteomics – Profiling the proteins UPEC produce inside the bladder and comparing them to defined lab conditions, revealing how real-life stressors and cues shape their physiology.
- Strain characterization – Isolating UPEC strains from patients and examining them in depth at both the genomic and phenotypic level to uncover traits linked to infection, persistence, and recurrence.
By combining these data sets, we are building a detailed picture of UPEC’s “lifestyle” inside the human urinary tract, knowledge essential for designing smarter, more effective therapies (manuscript in preparation).
Synthetic human urine (qUrine) for high-throughput UTI research
The behavior of bacteria strongly depends on their environment, including their susceptibility to antibiotics. In fact, the concentration of an antibiotic required to inhibit bacterial growth can vary up to 10,000-fold depending on whether the experiment is performed under standard laboratory conditions or in human urine.
However, working with real human urine poses several challenges. A major limitation is the lack of standardization, as urine composition varies from person to person and even from day to day, making it difficult to run reproducible lab experiments.
To address this, we created qUrine (quantitative urine), a fully synthetic medium that reproduces the essential features of real urine (in collaboration with Sebastian Hiller, Biozentrum, and Uwe Sauer, ETH Zurich, manuscript in preparation). The qUrine formulation captures key characteristics such as the high osmolarity typical of urine, the common nutrient composition, and the specific metabolic limitations, which can be leveraged for antimicrobial drug development.
In essence, qUrine provides a consistent, controllable environment for studying UPEC growth, adaptation, and treatments responses under realistic bladder-like conditions in a high-throughput compatible manner. It is now used both in our lab and by several other research groups worldwide to accelerate discoveries and enable cross-study comparisons.
Dynamic human bladder microtissue model for infection and treatment studies
Understanding urinary tract infections (UTIs) requires experimental models that capture the key features of the human bladder. In collaboration with the bioengineering group of Andreas Hierlemann (D-BSSE, ETH), we have developed a modular platform that combines a fully stratified 3D human bladder microtissue with a custom microfluidic device that mimics natural urine flow.
This setup allows us to follow how UPEC infections unfold in real time and at unprecedented resolution, from initial attachment, biofilm formation, and tissue invasion to the development of intracellular bacterial communities, and to observe how infections respond to treatment at single cells level (for both bacteria and the host tissue). We use this model to investigate fundamental pathogen–host interaction mechanisms and to evaluate therapeutic strategies, including standard antibiotics, novel antimicrobial agents (in collaboration with Rolf Müller, HIPS), pathoblockers (anti-virulence agents), and bacteriophage therapy (manuscript in preparation).
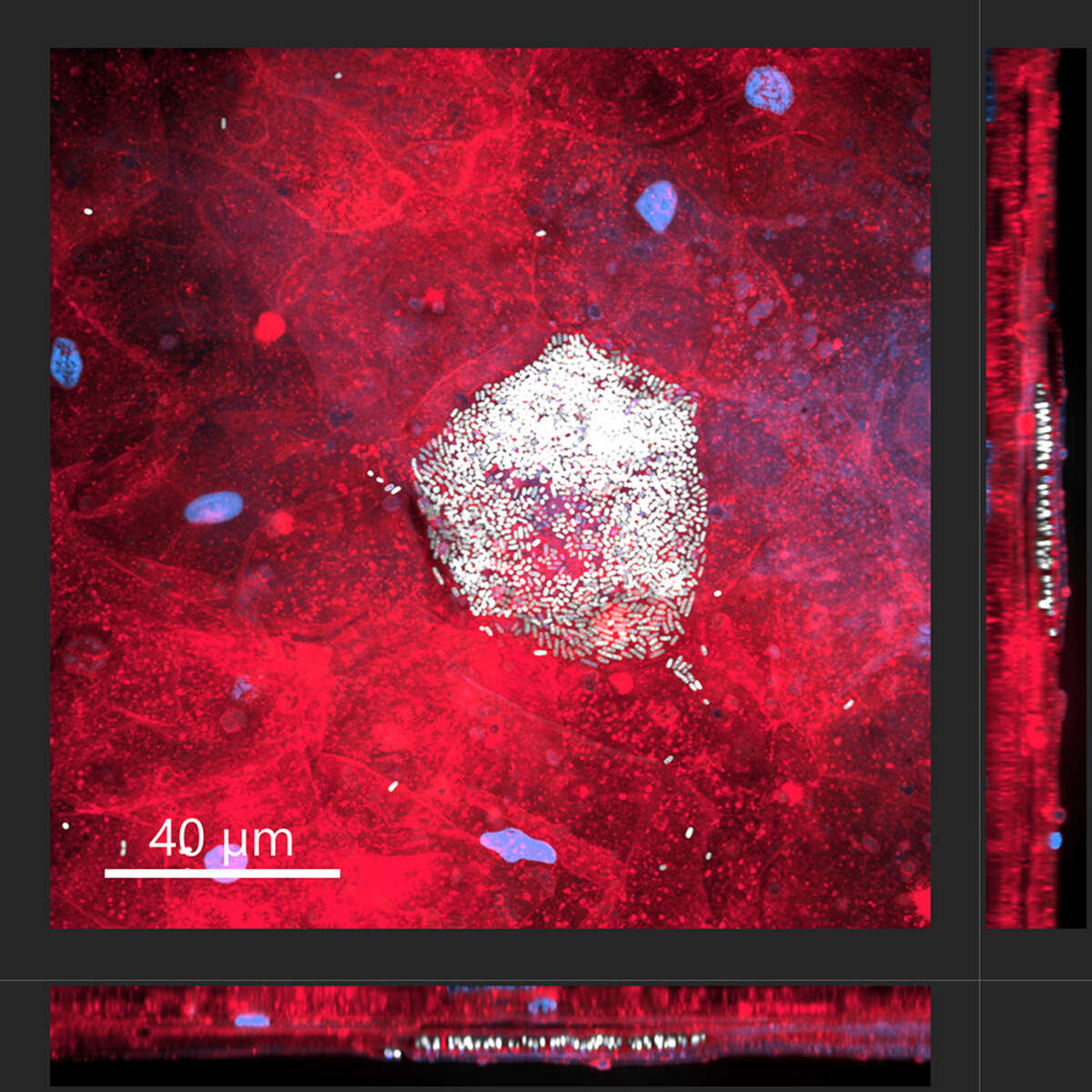
Intracellular bacterial community of uropathogenic Escherichia coli within an umbrella cells of a human bladder microtissue. Top view (large square) and side views (right and bottom rectangles) of a differentiated human urothelial microtissue, with eukaryotic cell membranes shown in red, nuclei in blue, and bacteria in white.
Recognizing that other pathogens also cause UTIs, we have extended the use of this model to Pseudomonas aeruginosa, a difficult to treat uropathogen responsible for up to 10% of complicated UTIs (in collaboration with Mike Givskov, Costerton Biofilm Center; Alexander Titz, HIPS).
An important aspect of our ongoing work focuses on adding immune competence to the model, enabling the study of the role of immune cells, such as macrophages and neutrophils, in the infection process (in collaboration with Lauriane Cabon, IHB, Roche; Kathin Bausch, University Hospital Basel, Thomas Kessler, Balgrist University Hospital).
By recreating key aspects of the human urinary tract, this platform bridges the gap between laboratory studies and real-world infections, providing a powerful tool to unravel UTIs pathogenesis and accelerate the development of more effective therapies.
CRISPRi Screening to uncover UPEC adaptation in the urinary tract
During UTIs, UPEC must adapt to a challenging environment, coping with the high osmolarity and low nutrient content of urine, withstanding shear forces from urine flow, evading attacks from immune cells such as neutrophils and macrophages, and successfully colonizing and invading the bladder lining.
To understand these adaptations at the molecular level, we need to identify the bacterial factors involved, some of which could serve as novel therapeutic targets. We use genome-wide CRISPR interference (CRISPRi) screening (in collaboration with David Bikard, Institut Pasteur) to pinpoint these factors. Our current work focuses on metabolic adaptations (manuscript in preparation), and we are expanding this research to explore stress responses and mechanisms that enable interaction with and invasion of the urothelium.
Brucella spp. and Brucellosis
Revisiting effector protein secretion in Brucella - host cell interactions
A hallmark of Brucella’s life cycle is its ability to survive and replicate within the endoplasmic reticulum (ER) of host cells. This depends on its unusual Type IV secretion system (T4SS), which delivers effector proteins into host cells to manipulate their functions and evade immune defenses. Despite its importance for virulence, the full set of Brucella T4SS effectors and the details of their secretion—distinct from canonical VirB-like systems—remain unclear.
Our previous work on the related pathogen Bartonella identified novel T4SS effectors, defined their secretion signals, and revealed how they are delivered into host cells. In Brucella, we use—further to the major human-pathogenic pathogens Brucella melitensis and Brucella abortus (both biosafety risk group 3)—also Brucella microti (a biosafety risk group 2 organism) to investigate T4SS function and effector secretion. We discovered that some putative effectors are periplasmic proteins released via outer membrane vesicles (OMVs, manuscript in preparation) and identified BspD, a conserved Rhizobiales effector involved in envelope integrity but not obviously coevolving with the VirB T4SS (Ketterer et al., 2024, mSphere). These findings suggest that Brucella’s effector landscape is more complex than previously thought.
In this project, we are using advanced tools — including bacterial secretome profiling with cell-selective metabolic labeling (BONCAT) and innovative translocation assays—to confirm known effectors, discover new ones, and define their secretion signals through structure-function studies (in collaboration with Xavier de Bolle, University of Namur). This work will reshape our understanding of how Brucella deploys its effectors, whether via the VirB T4SS or alternative pathways like OMVs, and reveal new aspects of its interaction with host cells.
Uncovering intracellular persistence and drug response of Brucella
We previously showed that Brucella abortus can persist inside host cells even after antibiotic treatment, retaining its ability to cause infection (Mode, Ketterer, Québatte et al., 2022, PLoS Negl Trop Dis). This finding pointed to an intracellular reservoir that may drive reinfection and relapse. Building on this, we are now using the same model for drug screening, aiming to repurpose existing compounds for safer and more effective brucellosis treatment.
To explore this phenomenon in patients, we collaborated with Jacob Moran-Gilad (Ben-Gurion University of the Negev) to study Brucella melitensis in blood samples from individuals with brucellosis. From these insights, we developed a PBMC-based infection model using a dual-color reporter strain to track intracellular survival and metabolic activity after antibiotic exposure.
Our studies revealed that a subset of intracellular B. melitensis remains metabolically active and resumes growth once antibiotics are removed. These results support the idea that the intracellular niche shields Brucella from treatment, and they establish our PBMC-based model as a powerful platform for evaluating antibiotic efficacy and testing novel therapeutic strategies, including drug repurposing.
Main collaborators:
- Lauriane Cabon, Institute of Human Biology, Roche, Basel, Switzerland
- Kathrin Bausch, University Hospital Basel, Basel, Switzerland
- David Bikard, Institut Pasteur, Paris, France
- Xavier de Bolle, University of Namur, Belgium
- Mike Givskov, Costerton Biofilm Center, Copenhaben, Denmark
- Andreas Hierlemann, D-BSSE, ETH Zurich, Basel, Switzerland
- Sebastian Hiller, Biozentrum, University of Basel, Basel, Switzerland
- Thomas Kessler, Balgrist University Hospital, Zurich, Switzerland
- Anders Meibom, EPFL, Lausanne, Switzerland
- Jacob Moran-Gilad, Ben-Gurion University of the Negev, Be'er Sheva, Israel
- Rolf Müller, HIPS, Saarbrücken, Germany
- Sven Panke, D-BSSE, ETH Zurich, Basel, Switzerland
- Uwe Sauer, ETH Zurich, Zurich Switzerland
- Alexsander Titz, HIPS, Saarbrücken, Germany
- Sarah Tschudin, University Hospital Basel, Basel, Switzerland
Main funding sources:
- NCCR AntiResist (phase II – 2024-2028): https://www.nccr-antiresist.ch/; https://data.snf.ch/grants/grant/225154
- SNSF project grant: Revisiting effector protein secretion in Brucella - host cell interactions (2024-2028): https://data.snf.ch/grants/grant/10003225
- Biozentrum, University of Basel


