Main Content
Support for Structural Bioinformatics
Computational structural biology is an increasingly important multidisciplinary field that uses advanced models and computational techniques to understand and solve complex problems concerning the structure and function of biological macromolecules. We combine a sound understanding of structural biology and biophysics with programming and software engineering competence.
In collaboration with the research groups involved in structural biology and biophysics research, we develop methods and scientific software to analyze and model structural, kinetic, as well as other types of data. Our support starts with open-door consulting and continues into long-term scientific and development projects. Some questions require highly specific code that is custom-built to address singular experiments. Wherever possible, however, we work on sustainable software solutions, applicable to a wider range of scientific questions.
Research and Development Highlights:
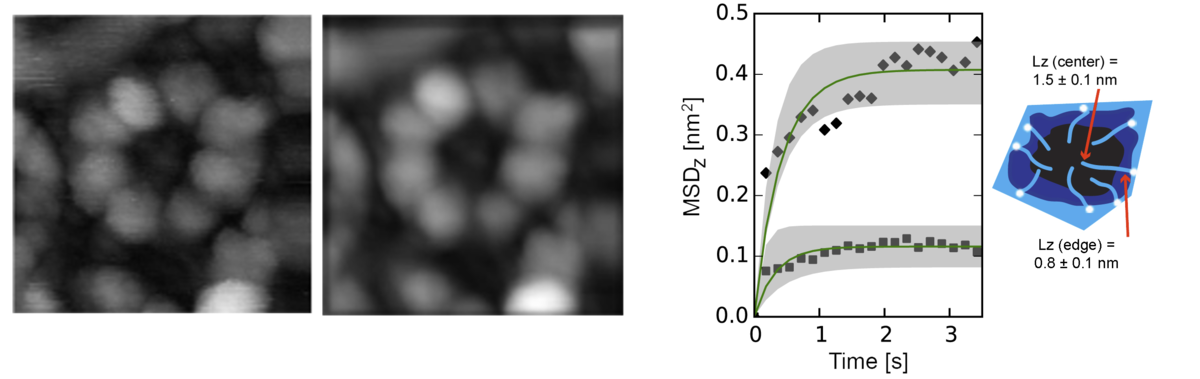
High-Speed Atomic Force Microscopy (HS-AFM)
This technique allows the direct observation of biomolecules in function at a high spatiotemporal resolution. We have developed a software package to process and analyze raw data, simulate AFM topographs and infer spatial and time-dependent properties from HS-AFM data.
For more information, see Nature Nanotechnol. 2016, 11(8):719-23 and ACS Nano. 2017, 11(11):10852-10859
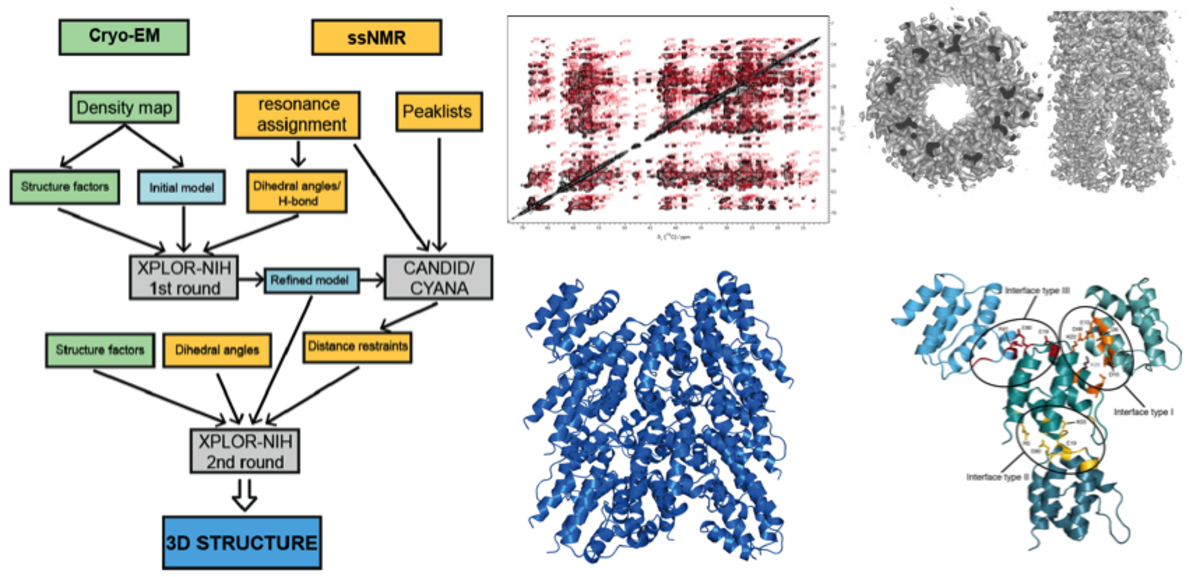
Integrative Structural Biology
We designed a protocol for structure determination that combines solid-state NMR and CryoEM experimental data simultaneously. This novel protocol was used to determine the structure of the mouse ASC-PYD (pyrin domain of apoptosis-associated speck-like protein) filament at atomic resolution.
For more information, see PNAS 2015, 112(43): 13237–13242 and Biomol. NMR Assign. 2016, 10(1):107-15
Pulse Program Database
We are currently developing a web platform for NMR pulse programs. This platform allows the archival and annotation of programs, as well as their deployment to an NMR instrument. While this tool is currently used internally, our goal is to provide the wider NMR community with a set of reliable, easy to use and well-annotated pulse programs.
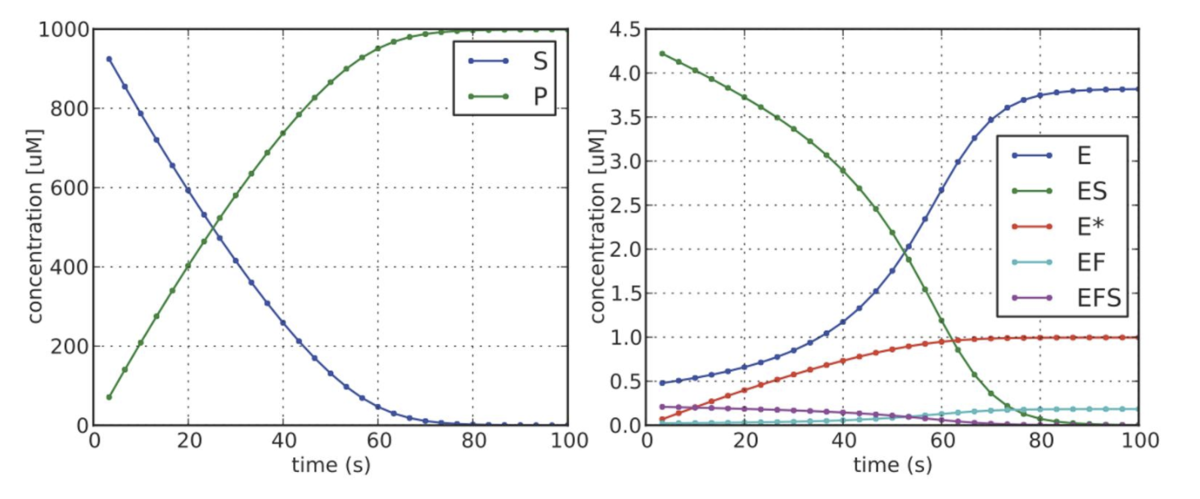
Kinetic data analysis
An appropriate experimental design and the use of proper statistical tools are prerequisites for the accurate analysis of enzyme kinetic data. We have developed an extensible simulation and data-fitting software for kinetic experiments in Python.
For more information, see PNAS 2016, 113(5):E529-37 and J. Bacteriol. 2015, 198(3):448-62