Main Content
Stochastic promoter choice during differentiation of neurons
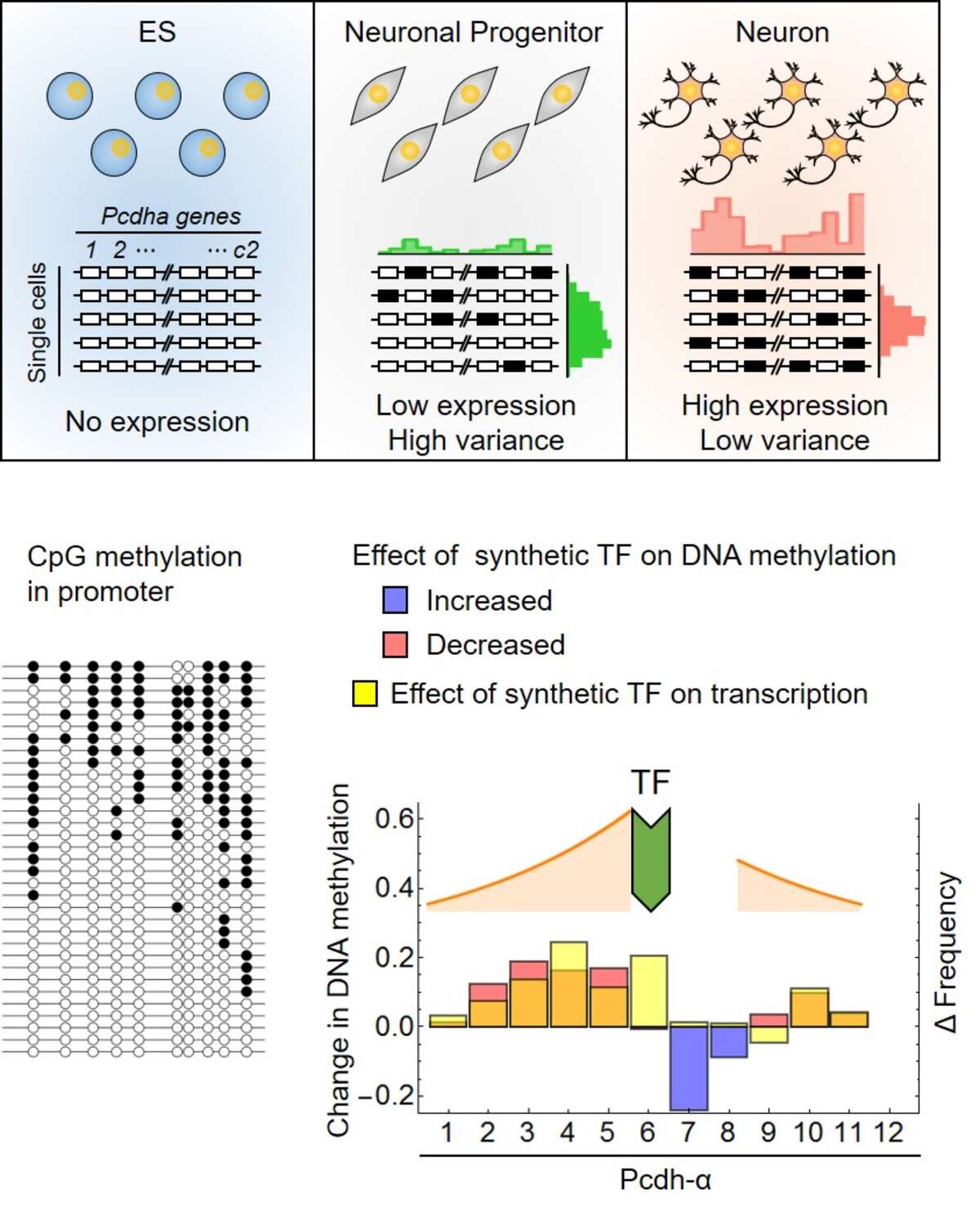
Mammals, including humans, have relatively few genes (20-50’000) but millions of neurons with distinct cellular identities. The combinatorial expression of protein isoforms is a strategy that can be used to create a diverse population of neurons with a limited number of genes.
Such combinations can arise by the random selection of gene isoforms, exemplified by the protocadherin (Pcdh) cluster of genes. At the same time, this randomness can lead to large fluctuations in the number of expressed isoforms in the individual cells, which can impair their functionality. By studying the stochastic combinatorial expression of the Pcdh array as neurons developed from embryonic stem cells, we showed with the help of a mathematical model that combinatorial diversity and precision are not mutually exclusive, but go hand in hand. The number of different isoforms in a cell and the exclusive precision during nerve cell maturation increase simultaneously.
T. Wada, S. Wallerich, A. Becskei, Stochastic Gene Choice during Cellular Differentiation. Cell Rep 24, 3503-3512 (2018).
Bistability and cellular memory
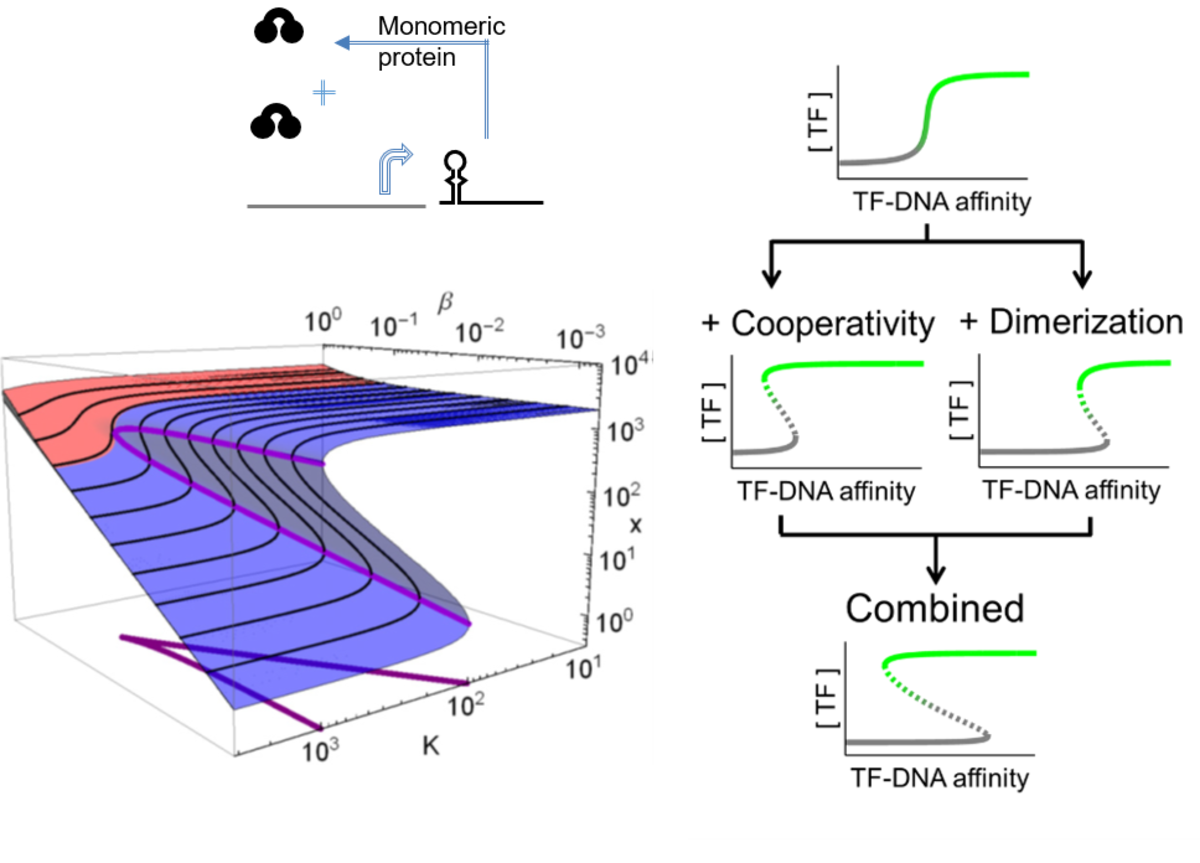
Even single cells are able to remember information. Cellular memory can be stored by positive feedback, which can generate two stable states, bistability, provided the loop incorporates nonlinear reactions. Despite the ubiquity of protein homodimerization, its ability to generate bistability remained elusive. We built synthetic feedback loops in yeast and modulated two nonlinear reactions: homodimerization and the cooperative binding of the transcription factor to the promoter. Either of them was sufficient to generate bistability on its own and when acting together, a particularly robust bistability emerged. Our finding was remarkable, since cells with a single-gene feedback loop incorporating two ultrasensitive reactions remembered the initial trigger even after 100 cell generations.
C. Hsu, V. Jaquet, M. Gencoglu, A. Becskei, Protein Dimerization Generates Bistability in Positive Feedback Loops. Cell Rep 16, 1204-1210 (2016).
I. Majer, A. Hajihosseini, A. Becskei, Identification of optimal parameter combinations for the emergence of bistability. Physical Biology 12, 066011 (2015).
Kinetics of post-transcriptional processes
In our recent work, we designed a multiplexed version of the gene control (MGC) method and found that mRNA turnover in yeast is very rapid, with a median half-life of around 2 min, which is substantially less than the 10 - 25 min range found by most other studies using transcriptional inhibition or metabolic labelling to determine half-lives. This permits a very dynamic functioning of RNA regulatory networks.
A. Baudrimont et al., Multiplexed gene control reveals rapid mRNA turnover. Sci Adv 3, e1700006 (2017).