Main Content
Research projects
We investigate the physiology of bacterial pathogens (Salmonella, Pseudomonas, Staphylococcus) in infected rodent and human tissues, focusing on single-cell behavior and potential applications for novel anti-infectives.
Host mechanisms against intracellular Salmonella
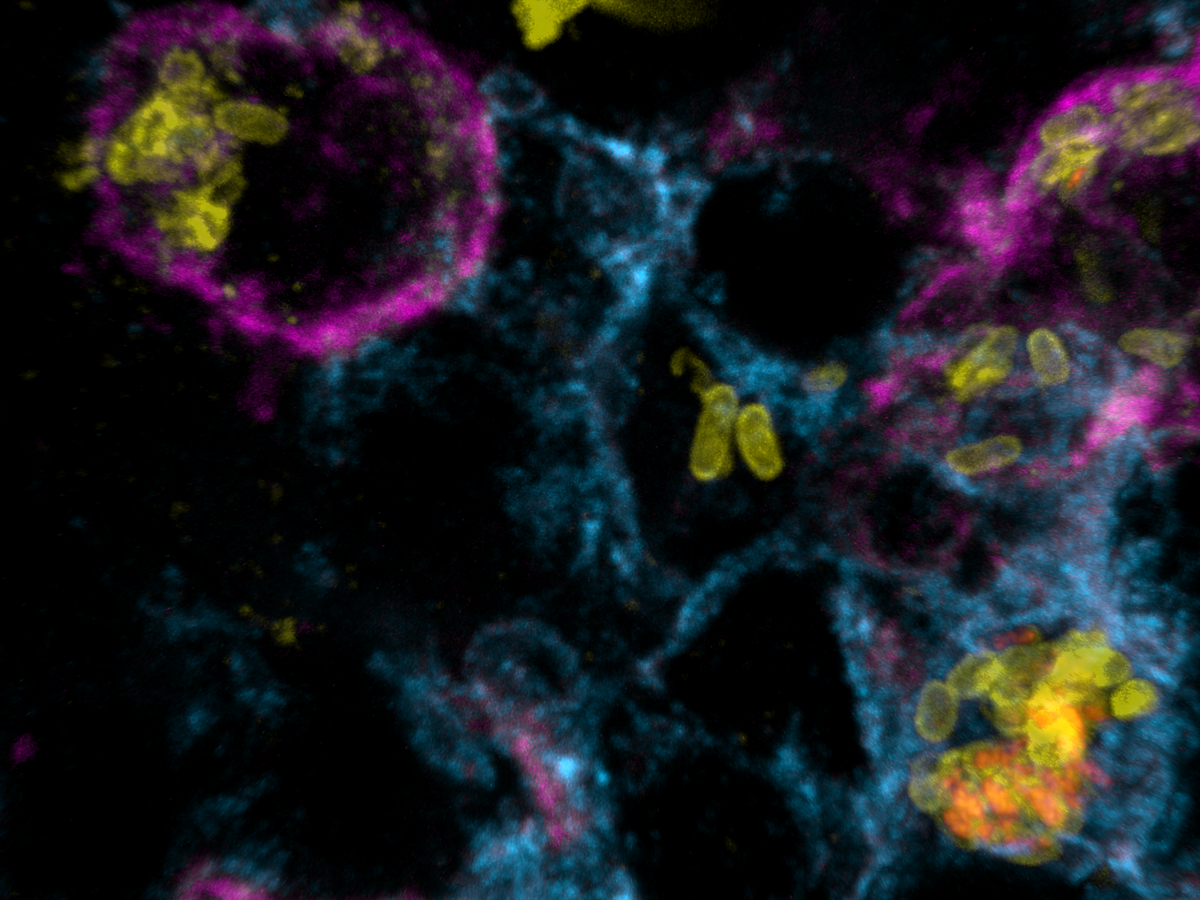
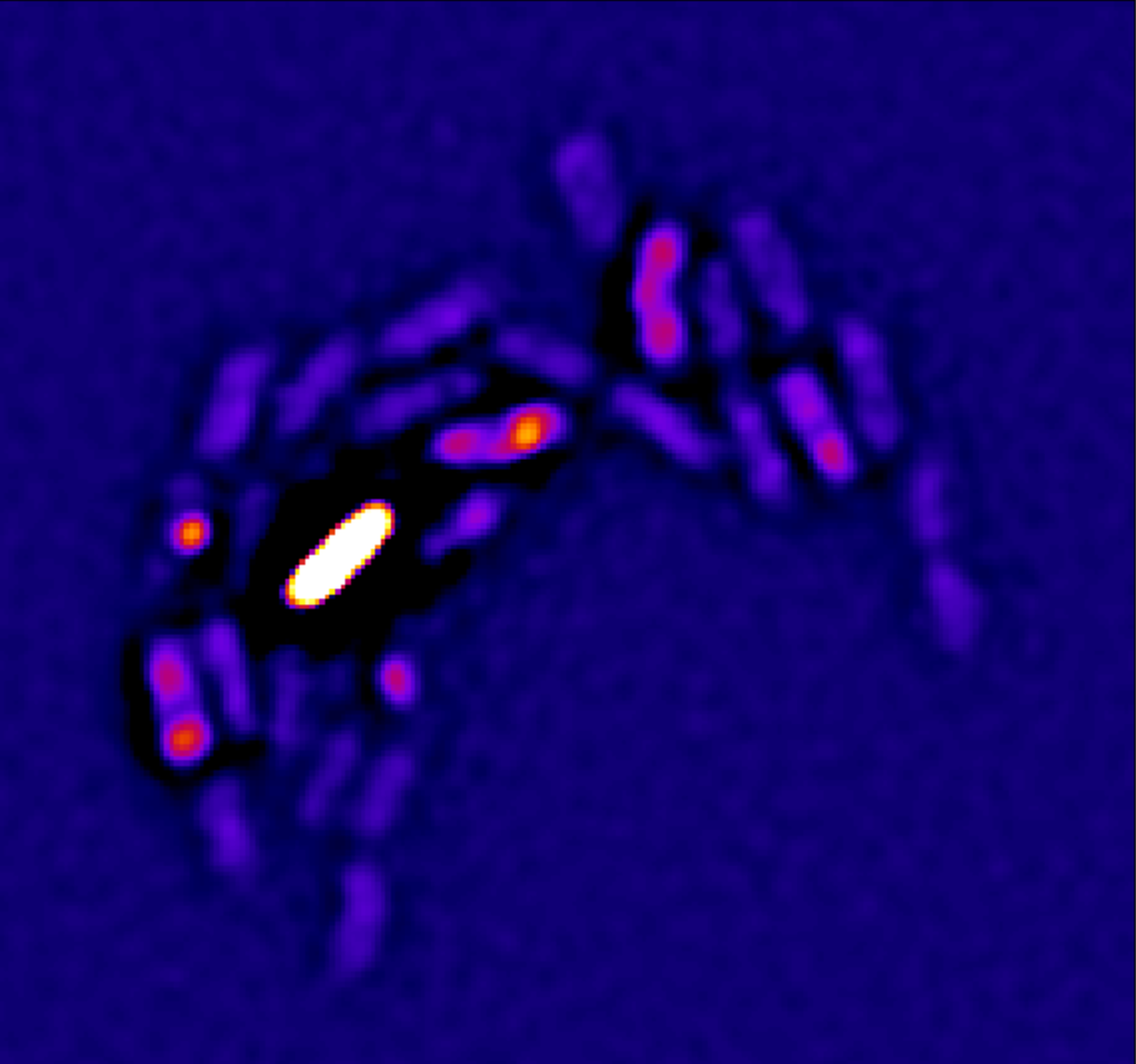
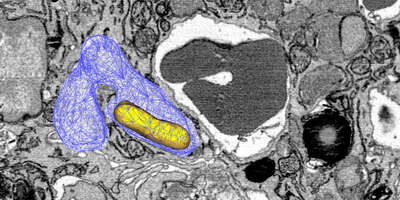
Salmonella causes diarrhea as well as severe systemic illnesses, such as typhoid fever. During systemic infections, Salmonella primarily resides within phagosomes inside host macrophages. We discovered that these Salmonella-containing vacuoles offer various nutrients, but only in limited amounts, resulting in slow bacterial replication. Magnesium is especially scarce, yet essential for bacterial physiology. In contrast, although iron is typically restricted by the host, Salmonella overcomes this limitation by targeting iron-rich vacuoles in macrophages that degrade old and damaged red blood cells.
Currently, we are investigating how magnesium limitation synergizes with envelope stress during infection.
Selected papers:
Roche, B., Claudi, B., Cunrath, O., Bleck, C., Antelo Varela, M., Li, J., Bumann, D. (2025). A Salmonella subset exploits erythrophagocytosis to subvert SLC11A1-imposed iron deprivation. Cell Host Microbe 33:632-642.e4. doi: 10.1016/j.chom.2025.04.013.
Cunrath O, Bumann D. (2019). Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 366:995-999.
Antibiotic action in vivo
Antibiotics clear Salmonella from infected tissues only slowly. Previously, this slow clearance was thought to result mainly from bacterial tolerance induced by stressful host conditions or from small, hyper-resilient bacterial subsets ("persisters"). However, we showed that Salmonella resilience in vivo is primarily due to their scarce nutrition, causing Salmonella to replicate slowly and become resilient against stresses including antibiotics.
Clearance of Salmonella is particularly slow in tissue compartments with weak local inflammation such as the white pulp of spleen. Boosting inflammation can support antibiotic clearance demonstrating the critical interaction of antibiotics with host immunity.
Furthermore, we discovered that standard plating assays commonly used to measure antibiotic killing can produce misleading results, falsely indicating the existence of hyper-resilient bacterial subsets. This problem arises because antibiotics remain bound to bacterial targets even after washing, causing substantial bacterial killing during subsequent regrowth on standard media. To overcome this limitation, bacterial killing must be assessed through single-cell, real-time analysis.
Currently, we extend our analysis to other antibiotics and anti-virulence strategies.
Selected papers:
Fanous J, Claudi B, Tripathi V, Li J, Goormaghtigh F, Bumann D. (2025). Limited impact of Salmonella stress and persisters on antibiotic clearance. Nature 639: 181–189.
Li J, Claudi B, Fanous J, Chicherova N, Cianfanelli FR, Campbell RAA, Bumann D. (2021) Tissue compartmentalization enables Salmonella persistence during chemotherapy. Proc Natl Acad Sci USA 118(51):e2113951118.
Human infections
Bacterial infections represent a major threat to global human health, yet we know very little about what actually occurs inside infected human tissues. Key questions remain unanswered: Where exactly do bacteria reside in human tissues? How do they respond to stresses and opportunities within the host environment? And how diverse are bacterial populations during infection?
Addressing these gaps is critical, as it directly influences the development and validation of relevant preclinical and in vitro models needed to identify and develop effective novel treatments. As part of the Swiss NCCR AntiResist network, we are extending our in vivo analyses from rodent models to the more challenging examination of human biopsies. These biopsies are collected through collaborations with the University Hospitals of Basel and Zurich from patients infected with Staphylococcus aureus, Pseudomonas aeruginosa, and E. coli.
In collaboration with AntiResist researchers, we have developed a novel fluorescent dye for detecting S. aureus in human biopsies, as well as an ultrasensitive mass spectrometry technique to accurately identify bacterial proteins despite the overwhelming presence of human proteins. Using these advanced tools, we perform high-resolution imaging and proteome analysis of biopsies, generating data that allow us to develop and validate laboratory models closely mimicking human infections.
Selected papers:
Antelo-Varela M, Bumann D, Schmidt A. (2024). Optimizing SureQuant for Targeted Peptide Quantification: a Technical Comparison with PRM and SWATH-MS Methods, Anal Chem 96:18061–18069.
Jantarug K, Tripathi V, Morin B, Iizuka A, Kuehl R, Morgenstern M, Clauss M, Khanna N, Bumann D, Rivera-Fuentes P. (2024). A Far-Red Fluorescent Probe to Visualize Gram-Positive Bacteria in Patient Samples, ACS Infect Dis 10(5):1545-1551. DOI: 10.1021/acsinfecdis.4c00060 (2024).
Ude J, Tripathi V, Buyck JM, Söderholm S, Cunrath O, Fanous J, Claudi B, Egli A, Schleberger C, Hiller S, Bumann D. (2021) Outer membrane permeability: Antimicrobials and diverse nutrients bypass porins in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 118(31):e2107644118.