Main Content
Introduction
Cell growth is highly regulated. Cells respond to nutrients or other appropriate growth stimuli by up-regulating macromolecular synthesis and thereby increasing in size. Conversely, cells respond to nutrient limitation or other types of stress by down-regulating macromolecular synthesis and enhancing turnover of excess mass. Thus, the control of cell growth involves balancing positive regulation of anabolic processes with negative regulation of catabolic processes. Growth is also controlled relative to cell division. In proliferating cells, growth is linked to the cell cycle such that most cells precisely double their mass before dividing. In other physiological contexts, such as load-induced muscle hypertrophy or growth factor-induced neuronal growth, cell growth is controlled independently of the cell cycle. Furthermore, in addition to the temporal control of cell growth described above, cell growth can be subject to spatial constraints. For example, budding yeast and neurons grow in a polarized manner as a result of new mass being laid down only at one end of the cell. Finally, in multicellular organisms, growth of individual cells is controlled relative to overall body growth such that the organs and tissues constituting the organism are properly proportioned.
The TOR signaling network
What are the mechanisms that mediate and integrate the many parameters of cell growth? In other words, what determines that a cell grows only at the right time and at the right place? Remarkably, the study of these mechanisms has been largely neglected, despite their clinical relevance and despite cell growth being, along with cell division and cell death, one of the most fundamental (and obvious) features of life. Also remarkable is the finding that cell growth control, regardless of eukaryotic organism or physiological context, seems always to involve the protein kinase TOR (Target Of Rapamycin) and its signaling network. TOR has thus become known as a central controller of cell growth. Indeed, the discovery of TOR led to a fundamental change in how one thinks of cell growth. It is not a spontaneous process that just happens when building blocks (nutrients) are available, but rather a highly regulated, plastic process controlled by TOR-dependent signaling pathways.
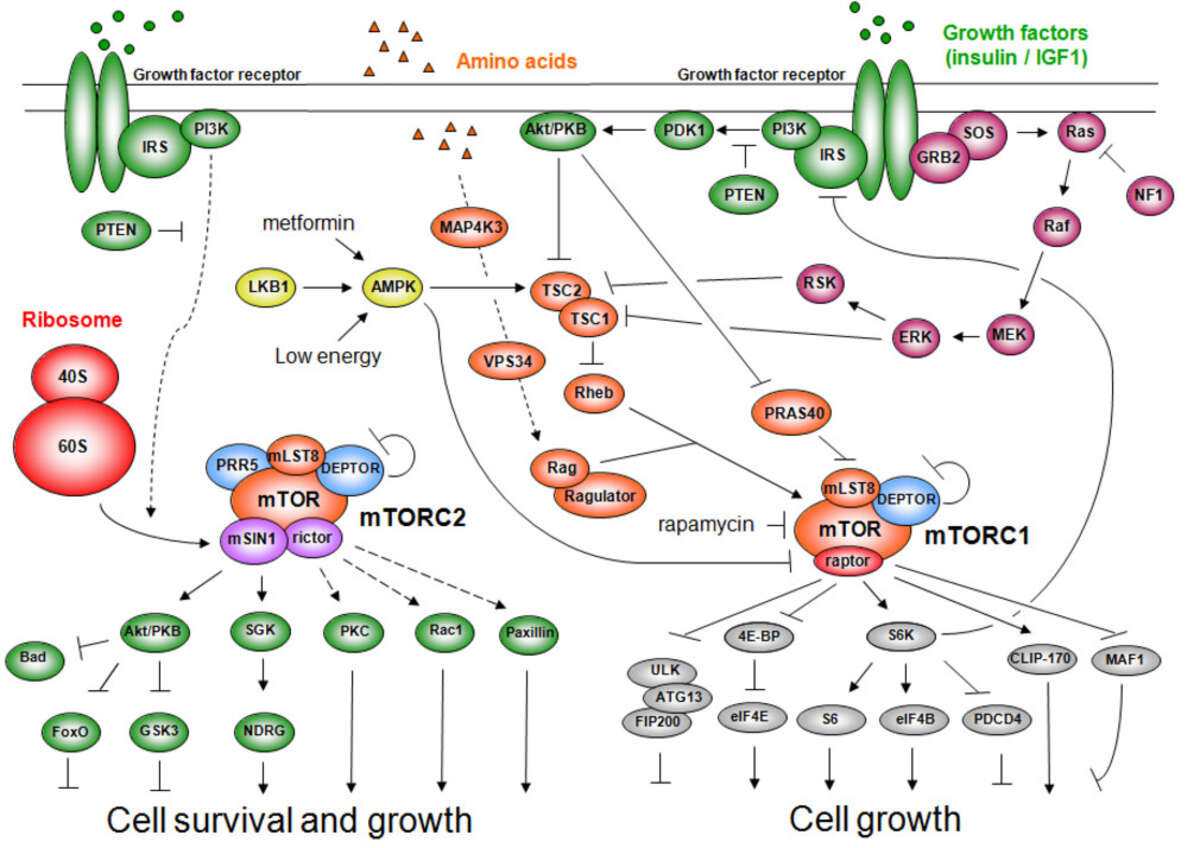
TOR, originally discovered in our laboratory, is structurally and functionally conserved from yeast to human (including worms, flies, and plants). TOR in mammals (mTOR) controls cell growth and metabolism in response to nutrients (e.g., amino acids), growth factors (e.g., insulin, IGF-1, PDGF), and cellular energy status (ATP). Nutrients are the dominant TOR input as high levels of amino acids can compensate for an absence of the other mTOR inputs but not vice versa, and only nutrients activate TOR in unicellular organisms. The growth factor signaling pathway, grafted onto the more ancestral nutrient sensitive TOR pathway, co-evolved with multicellularity. TOR activates cell growth by positively and negatively regulating several anabolic and catabolic process, respectively, that collectively determine mass accumulation and thus cell size. The anabolic processes include transcription, protein synthesis, ribosome biogenesis, nutrient transport, and mitochondrial metabolism. Conversely, TOR negatively regulates catabolic processes such as mRNA degradation, ubiquitin-dependent proteolysis, autophagy and apoptosis.
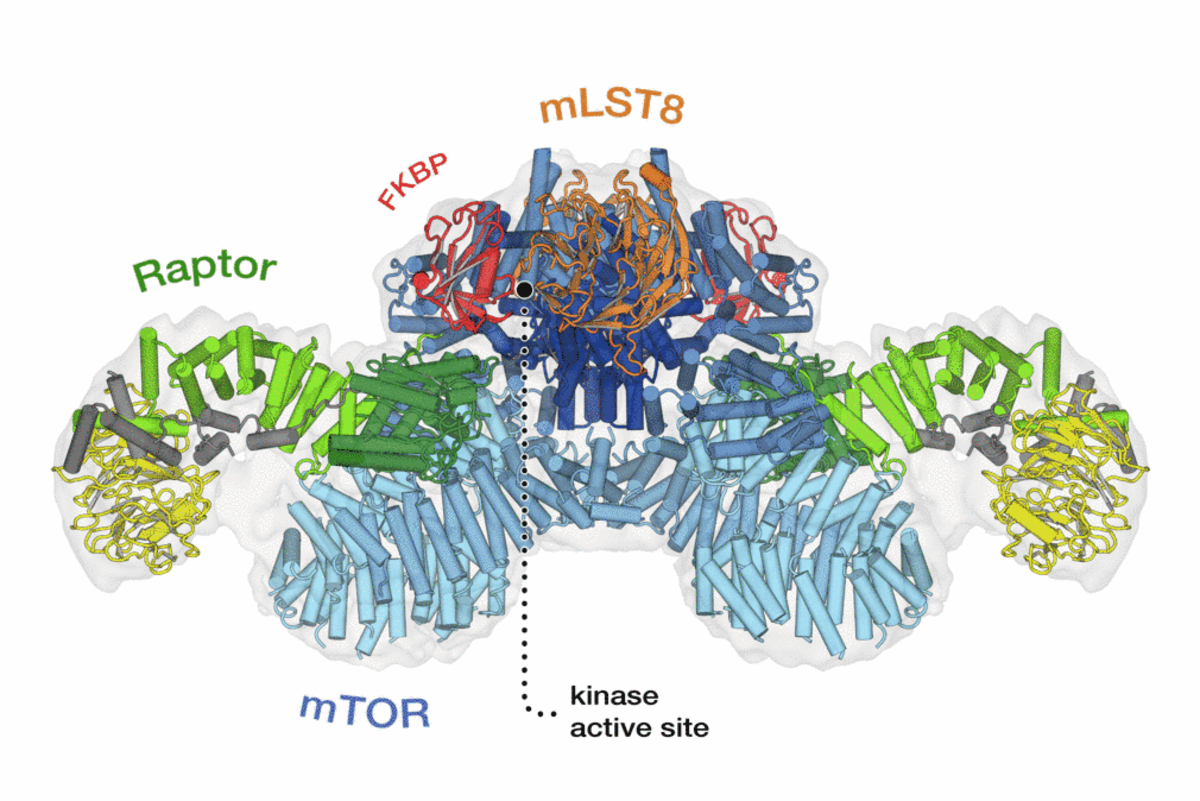
TOR is an atypical serine/threonine kinase that is found in two functionally and structurally distinct multiprotein complexes, TORC1 and TORC2 (mTORC1 and mTORC2 in mammals), each of which signals via a different set of effector pathways. TORC1 is rapamycin sensitive whereas TORC2 is rapamycin insensitive. The best-characterized phosphorylation substrates of mTOR are S6K and 4E-BP1 via which mTORC1 controls translation, and Akt/PKB via which mTORC2 controls cell survival and likely other processes. Like TOR itself, the two TOR complexes and the overall architecture of the TOR signaling network appear to be conserved from yeast to human. Thus, the TOR signaling network is a primordial or ancestral signaling network conserved throughout eukaryotic evolution to regulate the fundamental process of cell growth. As a central controller of cell growth and metabolism, TOR plays a key role in development and aging, and is implicated in disorders such as cancer, cardiovascular disease, obesity, and diabetes.
We are studying the TOR signaling network in the yeast Saccharomyces cerevisiae, in mammalian cells, in mice, and in human tumors. A major finding in our laboratory in recent years was the fact that TOR controls cell growth via two major signaling branches. Furthermore, we discovered the two TOR complexes and demonstrated that these two complexes correspond to the two previously described TOR signaling branches. More recently, in collaboration with our in-house colleague Markus Rüegg, we introduced the mouse as an experimental system to study the role of mTOR in regulating whole body growth and metabolism. In collaboration with the clinician Markus Heim, we have initiated a translational research project aimed at defining signaling pathways that allow tumors to evade therapy. The overall goal of our studies is to elucidate how growth and metabolism are regulated in health and disease.