Main Content
Neuronal circuit assembly in development and disease
We use a multidisciplinary approach to uncover fundamental mechanisms of neural development and plasticity. We combine advanced genetic, cell biological, electrophysiological, imaging, and computational methods to shed light on the mechanisms of synaptic wiring and the underpinnings of neurodevelopmental disorders.
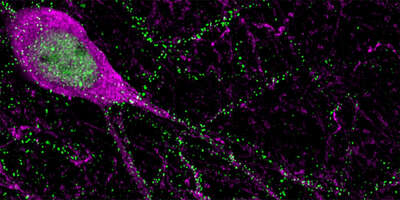
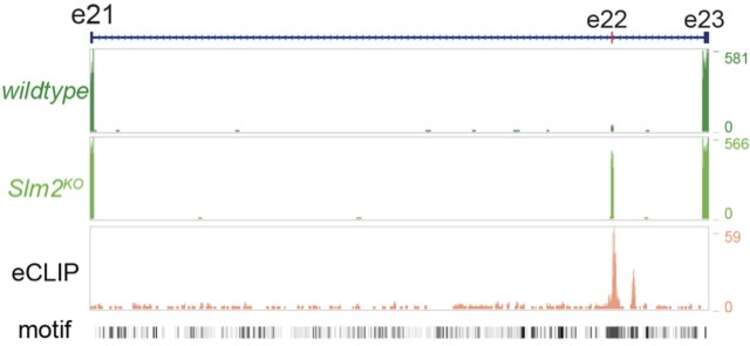
Alternative mRNA splice codes
We combine genetic, biochemical, and computational methods to uncover the fundamental contributions of post-transcriptional regulation (alternative splicing and translation initiation) to the developmental specification of synapses and the adaptation of circuit function to physiological states.
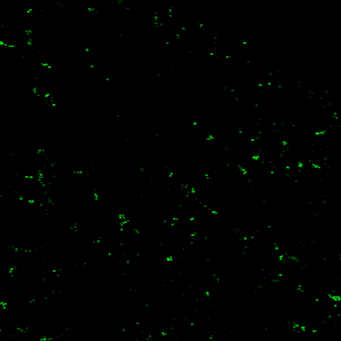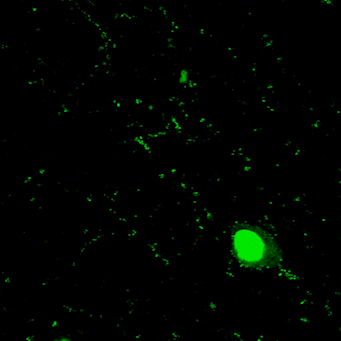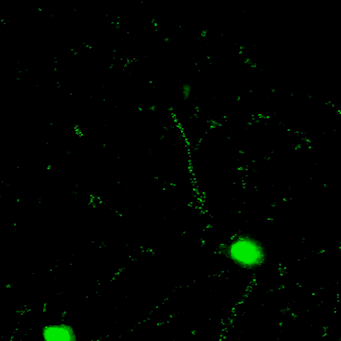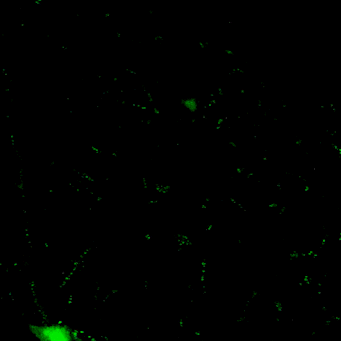
Novel tools for synapse marking
We are developing a powerful and versatile genetic tool-box for sparsely marking and quantitatively tracking synapse formation and neuronal function in the intact mouse brain. Key components are genetically-encoded fluorescent intracellular binders (chromobodies) and gene regulation elements (transposons, enhancers, splicing cassettes) to achieve spatio-temporal control for in vivo synapse biology studies in mice.
Spontaneous network activity
We use 2-photon in vivo imaging and genetic tools to record and manipulate activity of neuronal networks in newborn mice. By combining this approach with spatial transcriptomic methods, we are uncovering how spontaneous activity shapes cortical circuit development through molecular programs.


Social isolation, social reward, and autism-spectrum disorders
We are applying our multidisciplinary tool-set to understand the neural circuit underpinnings of social behaviors and their state-dependent adaptations. This work relies on opto- and chemogenetic approaches combined with deep-learning-based tools to quantify social interactions. A particular focus in this work is on neuropeptides and their receptors. The ultimate goal of these projects is the development of precision medicine approaches for autism.