Main Content
Research Projects
Biophysical Principles of Chaperone Function
Molecular chaperones play a key role in cellular processes, including protein homeostasis, but also in membrane protein transport and biogenesis. We employ high-resolution NMR studies of large chaperone-client complexes to provide an atomic resolution description of their structure and conformation and understand molecular mechanisms underlying their function.
Along these lines, we have used solution NMR spectroscopy to elucidate the mechanism underlying client recognition by the ATP-independent chaperones Spy, SurA and Skp at the atomic level (Burmann et al., Nat. Struct. Mol. Biol. (2013), Thoma et al., Nat. Struct. Mol. Biol. (2015), Morgado et al., Nat. Comm. (2017)). In these systems, we find that chaperones hold clients in a dynamic state that rapidly interconverts conformations while bound on the chaperone surface. Several chaperones were found to interact with the partially folded client Im7 by selective recognition of flexible, locally frustrated regions in a dynamic fashion (He et al., Sci. Adv. (2016), He et al., Angew. Chem. (2018)). By increasing client backbone dynamics, the chaperone facilitates the search for the native structure. The similarity of the interactions suggests that the underlying principle of recognizing frustrated segments is of fundamental nature (Hiller, Trends Biochem. Sci. (2019)). Chaperones thus resemble chaotropes in their action and there might be similarities in the underlying mechanisms (Macosek et al., Front. Mol. Biosci. (2021)).
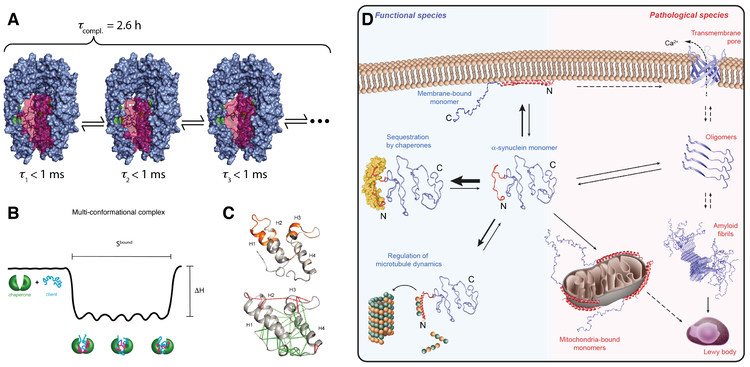
Recently, we have systematically investigated how molecular chaperones interact with the intrinsically disordered protein α-synuclein. α-synuclein plays a key role in Parkinson’s disease. We could show that molecular chaperones have a specific interaction signature and use this signature to elucidate how chaperones regulate the state of α-synuclein in living cells (Burmann et al., Nature (2020)). Our ongoing projects in the field of chaperone biophysics address fundamental questions. We want to derive quantitative predictors for chaperone function, unravel atomic-level details of how clients are recognized, understand how the interplay works between chaperone and client dynamics and study how basic chaperone function is embedded into complex functional cycles.
Outer Membrane Protein Biogenesis Targeted by Novel Antibiotics
Insertion of proteins into the bacterial outer membrane is mediated by a dedicated chaperone machinery. At least eight different chaperones, arranged in an assembly line, transport β-barrel outer membrane proteins (Omps) and fold them into the membrane. Thereby, functioning of the insertase complex BAM is most fascinating, both from the view of fundamental biophysics, as well as from the perspective of microbiology. We aim at unraveling the underlying insertion mechanisms at the atomic level, following our initial hypothesis of a hybrid barrel mechanism (Gruss et al., Nat. Struct. Mol. Biol. (2013)).
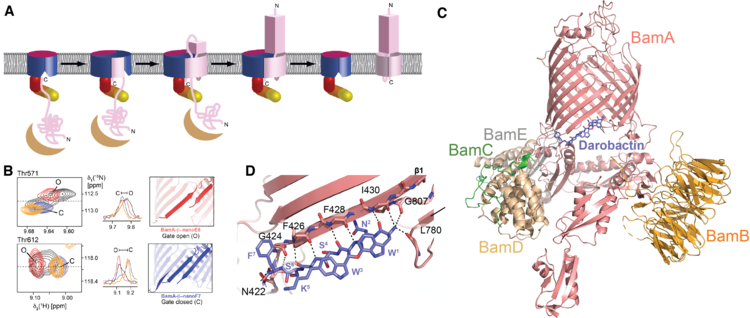
We have established NMR spectroscopy of the β-barrel BamA in detergent micelles, the largest integral membrane protein assigned to date (Hartmann et al., J. Am. Chem. Soc. (2018)). These NMR assignments have allowed to monitor the conformational ensemble at the atomic level and to map its interactions with two novel classes of antibiotics, darobactin and OMPTA, (Luther et al., Nature (2019); Imai et al., Nature (2019)). Darobactin binds the lateral gate of BamA in the BAM complex by β-strand augmentation. It mimics the natural β-signal sequence of substrates to block the entry of natural substrates (Kaur, Jakob et al., Nature (2021)).
The darobactin mode of action is unprecedented in structural biology. Open questions remain: how exactly does the transition from chaperone to BAM complex occur? How does the antibiotic OMPTA function? Can the BAM complex be inhibited by nanobodies?
Mechanisms of Bacterial Cell Signaling
Microorganisms can survive in challenging environments by rapidly adapting to the external conditions. Sensing external stimuli and transducing this information to the interior is in many cases mediated by phosphate signaling systems. Typical systems are composed of at least one sensor histidine kinase and one response regulator protein that communicate with each other via a phosphorylation cascade. In addition, Caulobacter and other bacteria use these systems to regulate their cell cycle. In such cases, not an external stimulus, but the fluctuating internal second messenger cyclic di-GMP is sensed.
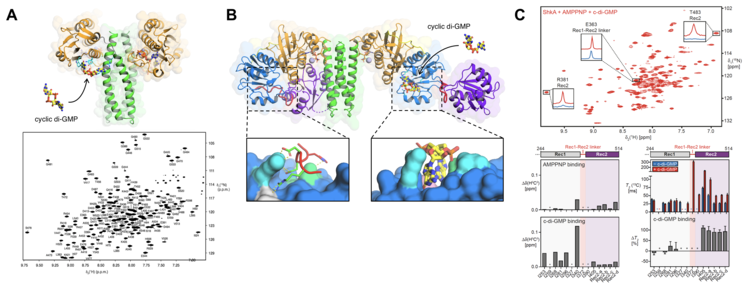
In close collaboration with the Jenal and the Schirmer labs, we use a combination of methods, in particular X-ray crystallography and NMR spectroscopy, to elucidate the underlying molecular mechanisms at the atomic level. We have shown that the activity of the histidine kinase CckA from the CckA-CtrA pathway involved in initiation of chromosome replication is regulated by cyclic di-GMP, we identified a novel cyclic di-GMP binding pocket in CckA and demonstrated that cyclic di-GMP reversibly locks two kinase core domains in place, hindering the hinge motions required for autophosphorylation (Lori et al., Nature (2015), Dubey et al., Sci. Adv., (2016)).
Recently, we focused on the ShkA-TacA pathway involved in the G1/S phase transition and stalk formation (Kaczmarczyk et al., Nat. Comm., (2020)). We demonstrated that cyclic di-GMP binds to the pseudo-receiver domain of the ShkA hybrid histidine kinase, thereby discovering a novel binding mode for cyclic di-GMP. Specific methyl-labeling NMR strategies on the full-length protein proved that the interaction of ShkA with cyclic di-GMP leads to a large conformational change in which the C-terminal domain is liberated from its obstructing position (Dubey et al., PNAS, (2019)).
We are currently interested in understanding how the proteins in more complex signaling transduction systems communicate with each other. Due to their large size, these proteins have not yet been much studied by NMR spectroscopy, therefore the functional dynamics are largely unknown. With modern NMR technology solutions are however well within technical reach. In addition, we aim at dissecting the mechanisms of response regulator-histidine kinase hybrids, which have shown to be crucial for virulence and motility in pathogenic bacterial species.
Mechanisms that Control Pyroptosis and Chronic Inflammation
The innate immune system provides the first line of defense against invading pathogens. Pathogen detection is dependent on the so-called pattern recognition receptors that recognize conserved structures of microbial origin. This leads to an inflammatory reaction in the host where a subset of the receptors, such as NLRP3, Pyrin, NLRC4 and AIM2, recruit ASC to assemble inflammasome complexes. The latter are multiprotein complexes that, upon assembly, drive caspase-1 activation leading to inflammation and pyroptosis, a pro-inflammatory form of cell death, mainly via activating gasdermin-D and release of pro-inflammatory cytokine interleukins, IL-1b and IL-18.
The macromolecular inflammasome complexes are of the order of 1-2 µM in diameter and typically a single complex, also called “speck”, is formed per cell. Inflammasome-induced IL-1b and IL-18 release is a physiological event necessary to fight infections, however, prolonged and uncontrolled activation of inflammasome complexes, especially by sterile inflammatory triggers, causes various (auto)-inflammatory and central nervous system (CNS)-related diseases. The main cellular receptor activated by sterile (auto)-inflammatory triggers is NLRP3 that in turn assembles the NLRP3-inflammasome. As such, inhibiting the NLRP3-inflammasome or pyroptosis in general holds promise as a therapeutic target for (auto)-immune diseases, as well as CNS-related diseases.
Over the last few years, we have determined the structure and assembly of the mouse ASC inflammasome (Sborgi et al., PNAS (2015)) and have demonstrated that the formation of the inflammasome speck correlates with receptor activation and is required for the production of IL-1b and IL-18 (Sborgi et al., PNAS (2015), Dick et al., Nature Communications (2016)). Moreover, we have also shown that the membrane pore-forming protein, gasdermin-D, plays a major role in pyroptosis and propagation of the inflammatory cascade via release of pro-inflammatory interleukins (Sborgi et al., EMBO Journal (2016), Heilig et al., Eur. J. Immunol. (2018)). Based on our data and available structural data on inflammasome receptors, we aim further to understand and manipulate how ASC-inflammasome and pore-forming gasdermin-D sustain chronic inflammation. As one approach, we are currently developing assays towards finding small molecules that inhibit pyroptosis and abate chronic inflammation (Sborgi et al., Cell Stress (2018)). By finding novel molecules that control inflammasome activation and pyroptosis, we aim to be at the interface of basic and applied research.
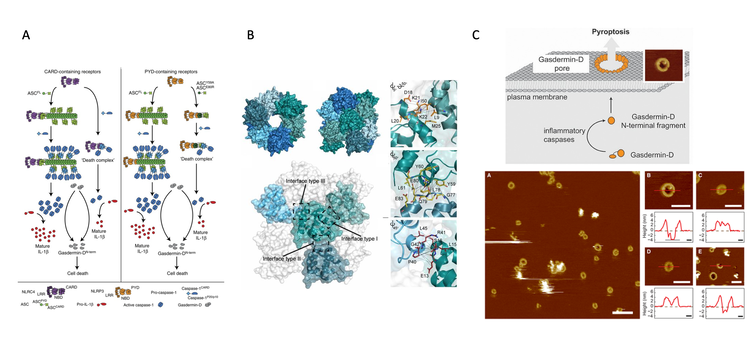
Figure 4: Mechanisms that control pyroptosis and chronic inflammation. (A) Model of signal amplification leading to pyroptosis induction by ASC filaments. (B) Top and side views of the ASC-PYD filament obtained by Cryo-EM. Four ASC-PYD monomers are shown in Surface representation showing four ASC-PYD monomers. The zoomed boxes show detailed interaction interfaces of the monomers. (C) Top – Model of GSDMD action in pore formation and pyroptosis induction. Bottom - AAFM topograph of GSDMDNterm bound to lipid membranes. (See respective publications for more detailed explanations)



![[Translate to Deutsch:] Model of chaperone droplet formation.](/fileadmin/_processed_/3/c/csm_News_Hiller_NatureCellBiolog_2025_4871e2e6a9.jpg)
