Main Content
Secretion systems allow bacteria to transport macromolecules such as proteins into host cells during pathogenesis or bacterial cells during competition in various ecological settings. Type 6 Secretion Systems (T6SS) are encoded by a cluster of 15-20 genes that is present in at least one copy in approximately 25% of all sequenced Gram-negative bacteria. Although linked to virulence during host infection, species such as Pseudomonas, Burkholderia, Acinetobacter and Vibrio can use T6SS to kill competing bacterial cells by delivery of toxic proteins in a cell-cell contact-dependent process.
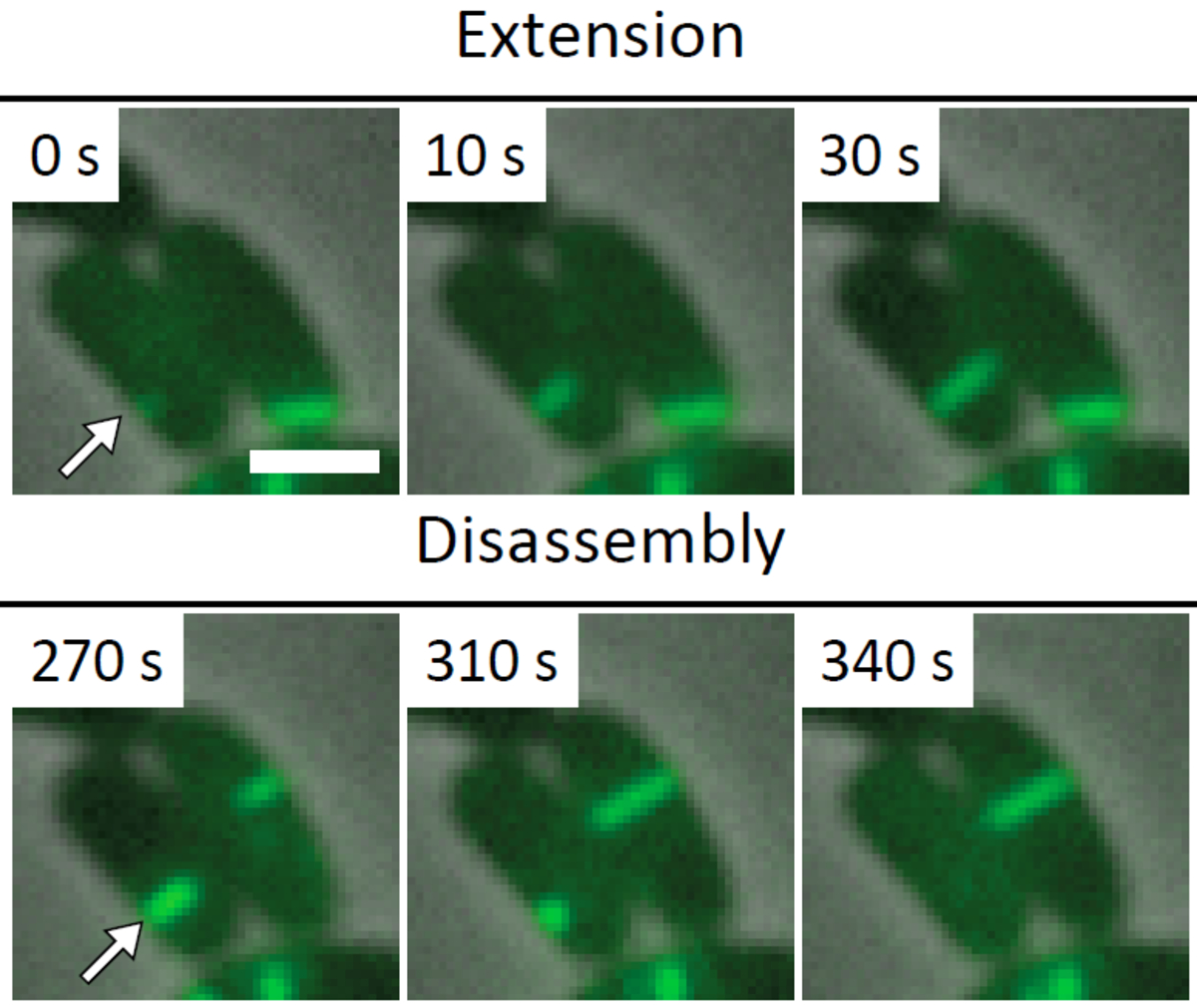
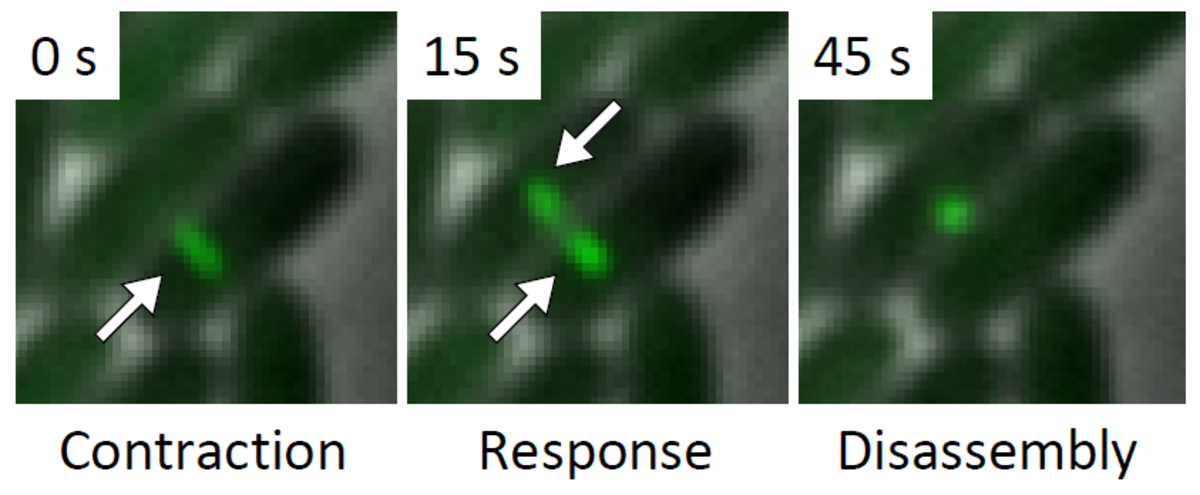
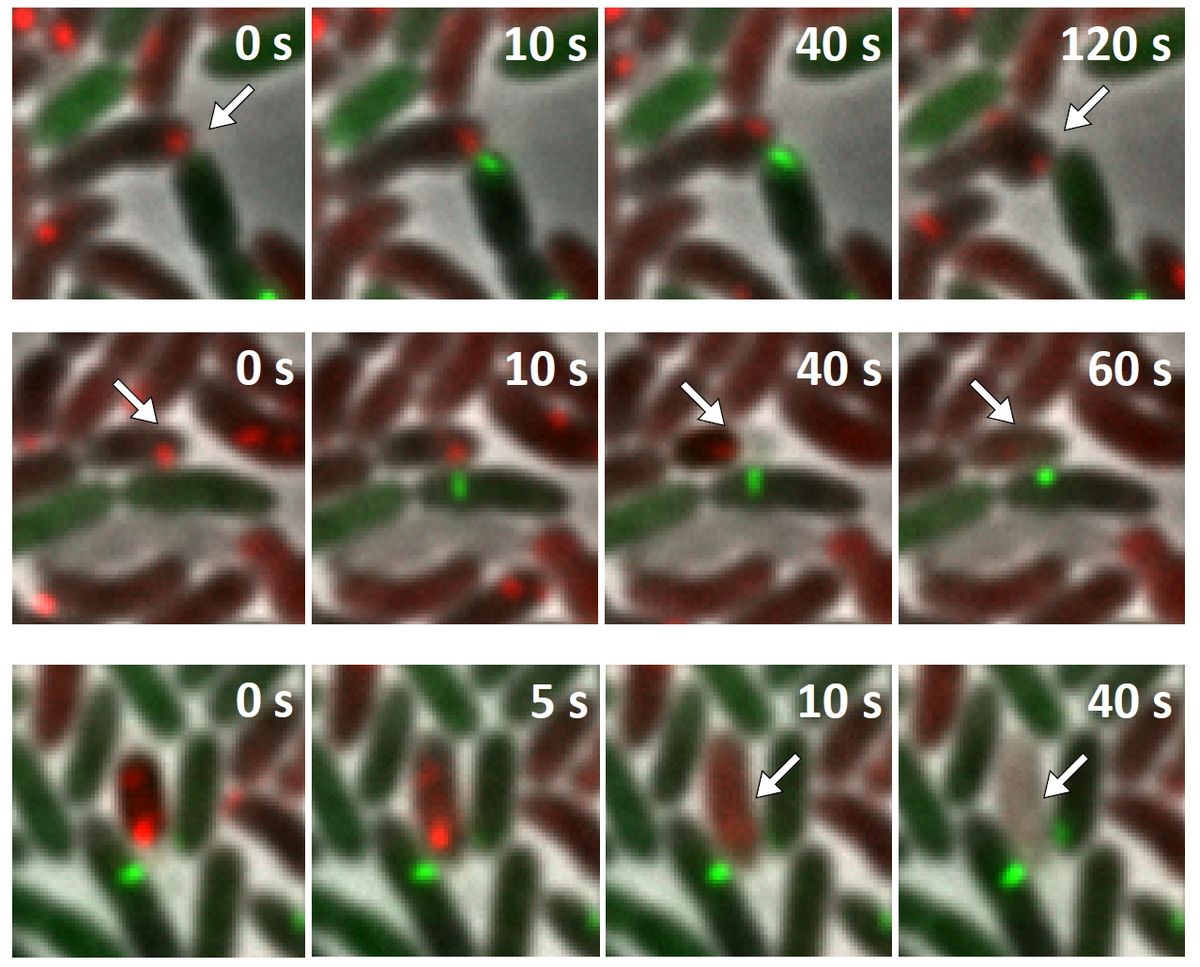
Several T6SS components are structural homologs of components of a contractile bacteriophage tail and assemble into a large structure that can be studied using whole cell electron cryo tomography and live cell fluorescence microscopy. A combination of these techniques allows to obtain high resolution structure of T6SS in situ and to follow T6SS assembly in time. This provides an unprecedented level of understanding of this dynamic nanomachine (Basler et al., Nature 2012, Basler and Mekalanos, Science 2012). For example, live cell imaging of T6SS activity in P. aeruginosa revealed that these cells are able to sense an attack from neighboring heterologous bacteria and assemble its T6SS apparatus with a remarkable precision to specifically kill an attacking cell without damaging bystander cells (Basler et al., Cell 2013).
The goal of our research is to understand the key molecular mechanisms underlying the assembly, substrate delivery, and regulation of T6SS function. We aim to create a detailed model of the T6SS nanomachine that would allow predicting structural changes needed to engineer T6SS with new properties. Novel approaches developed to study T6SS function will be further applied to study other molecular nanomachines as well as mechanisms of various contact dependent bacterial interactions in polymicrobial communities.
High-resolution structure of T6SS
A whole T6S apparatus was recently visualized in V. cholerae by whole cell cryo electron tomography. Resolution of the structure is, however, too low to identify individual components and therefore does not provide enough information to infer a mechanism of T6SS assembly. We are solving atomic resolution structures of T6SS components and analyze their mutual interactions by genetic and biochemical methods. We are developing novel strategies to improve resolution of the T6SS structure in situ in various model organisms. We aim to identify differences in T6SS assemblies in these organisms to explain the fundamental differences in their dynamics.
Visualization of T6SS activity
Our recent success in visualizing T6SS assembly and dynamics in live cells significantly improved our understanding of T6SS function in V. cholerae and P. aeruginosa. We use novel imaging approaches to describe localization of T6SS components with high spatial and temporal resolution. We are interested in understanding the process of initiation of T6SS assembly in various model organisms to further extend our knowledge about T6SS regulation. We are also using imaging to characterize the mode of action of T6SS effectors to better understand T6SS function.
Regulation of T6SS function
We are developing novel genetic methods to describe signaling processes involved in regulation of T6SS function on the transcriptional, translational, and post-translational level. We are also developing approaches to understand the role of T6SS in polymicrobial communities and to unravel the entire repertoire of secreted effectors.