Main Content
Unraveling the functioning and evolution of genome-wide regulatory networks
Using theoretical and experimental approaches to study the function and evolution of the regulatory networks that cells use to control the expression of their genes.
Gene regulatory networks underlie much of the characteristic complex behaviors of biological systems. They allow single-celled organisms to "act on their own behalf" by changing their internal state in response to changes in their environment, and they allow multi-cellular organisms to express a single genome into many different cellular phenotypes, allowing for "societies" of cells in which there is division of labor. We now know that gene regulatory networks are ultimately encoded by constellations of regulatory sites on the DNA that are recognized by regulatory proteins such as transcription factors (TFs) which "read out" the regulatory programs encoded in the genome.
Deciphering the regulatory code of the genome
In contrast to the large genes that encode proteins, the small regulatory sequences that control gene expression are generally hard to find. Moreover, little is known about their functioning. Our group develops mathematical and computational methods for deciphering this "regulatory code" in the DNA, and for modeling how constellations of regulatory sequences control gene expression.
Gene expression dynamics: from single cells to genome-wide regulatory programs
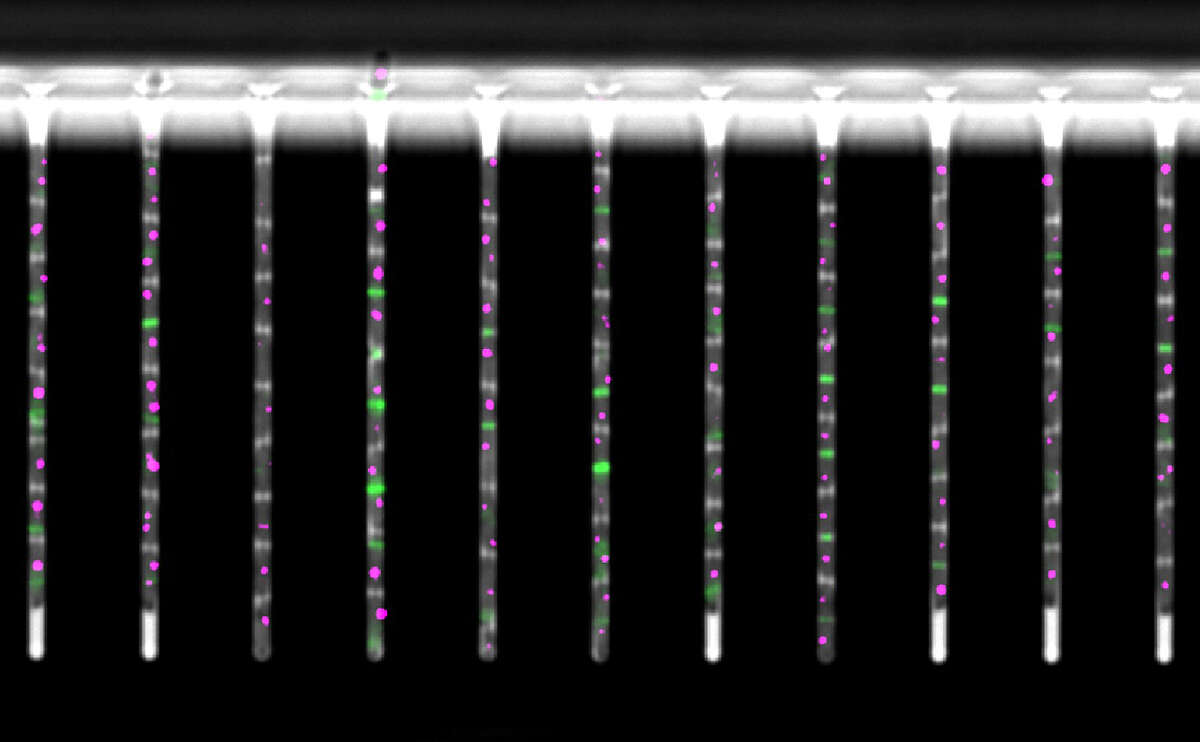
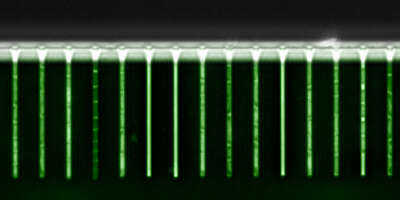
Recently developed experimental technologies make it possible to comprehensively monitor the gene expression and chromatin state of cells. We integrate computational predictions with such experimental datasets to model the dynamics of gene regulation, ranging from single-cell dynamics in E. coli, to genome-wide regulatory networks in human. Our ultimate goal is to not only understand but also learn how to actively manipulate the regulatory networks that control cellular behavior. Such ability has potential applications ranging from "domesticating" pathogenic organisms to engineering human tissues.
Discovering the laws of genome evolution
Darwin's theory of evolution by natural selection is the overarching theoretical framework for all of biology. However, despite almost a hundred years of evolutionary theory development, there are still virtually no real-world situations in which evolutionary theory can make concrete quantitative predictions. We believe that one crucial reason for this failure is that the key variables that feature in almost all mathematical population genetics models are fundamentally unmeasurable. With the advent of large-scale genomic sequencing it has become possible to empirically study quantitative patterns in genomic data and this has led to the discovery of a number of unexpected quantitative laws. Our group works on identifying such quantitative laws from large-scale genotype and phenotype data, with the aim of developing theories of evolution that are firmly rooted in directly measurable quantities.