Bacteria may be small, but they have a remarkable ability to adapt. They colonize a wide variety of habitats – from the human gut to hot springs – and respond with great flexibility to changes in their environment. One reason for their enormous adaptability is their ability to selectively turn on or off genes in response to external stimuli. The expression of genes is controlled by special regulatory proteins called transcription factors, which bind to specific DNA regions next to their target genes and thereby either suppress or enhance their activity.
Same signal, different response
Gene regulation in bacteria has often been described as a simple on/off switch – like flipping a light switch. The basic picture was that changes in the environment would cause transcription factors to either increase or decrease in concentration, and that this in turn would cause their targets to become more of less active. In their current study published in PRX Life, the team led by Prof. van Nimwegen reveals that in reality it is far more complex. Rather than being simply switched on or off, the concentrations of transcription factors can fluctuate vary rapidly and transiently, causing bursts of activation in their targets, and causing different targets to respond differently to the same stimulus, even when regulated by the same transcription factor.
Gene expression is unavoidably affected by molecular noise. As molecules are constantly moving and bouncing into each other, transcription factors do not bind to DNA continuously —but spontaneously bind to and unbind from the DNA. The fraction of time the DNA of a target gene is bound by a transcription factor depends on the concentration of this transcription factor and this in turn determines the expression level of the target.
How bacteria handle DNA damage
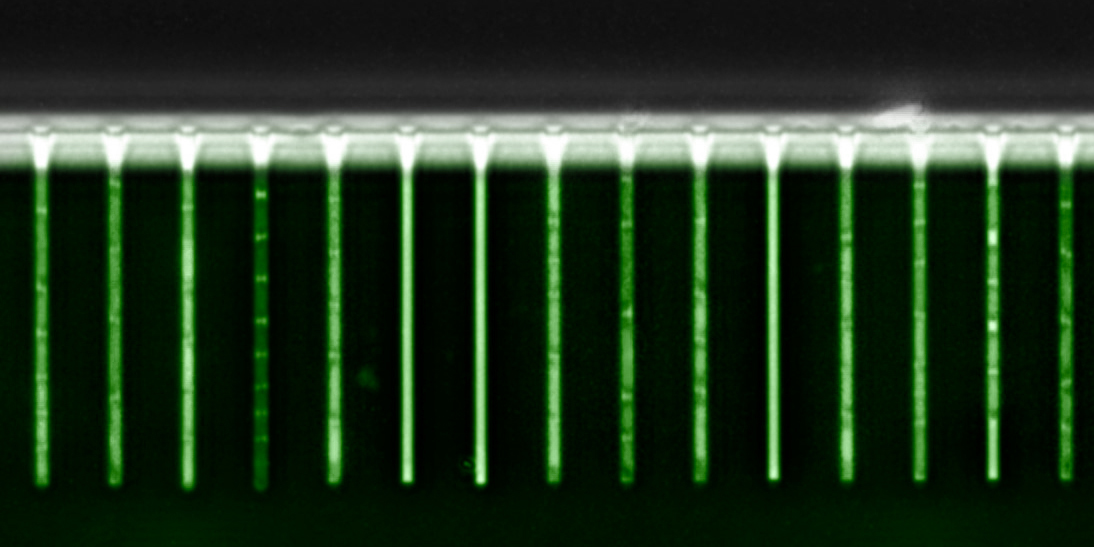
The researchers investigated DNA repair genes in E. coli which are regulated by the transcription factor LexA. Under normal conditions, the transcription factor LexA binds to DNA repair genes and prevents their transcription. DNA damage induces the cleavage of LexA, lowering its concentration and thus causing the repair genes to be bound less often and thereby induced. Using green fluorescent proteins, the researchers were able to observe in real time and in individual cells how the gene expression of different targets of LexA changes in response to DNA damage. In particular, by tracking changes in green fluorescence in single cells through time, the researchers could quantify precisely how strongly and for how long different genes were active in single cells.
Genes respond individually
Contrary to previous assumptions, the team found that genes were not activated in a continuous manner, but that each target gene exhibited short, random bursts. “When we induce DNA damage using an antibiotic, the cells show short burst of production of green fluorescence,” says van Nimwegen. “and these bursts have very specific and distinct characteristics for different LexA target genes.” Interestingly, although the short expression bursts become more frequent as the antibiotic concentration increases, the frequencies vary across target genes and so do their amplitudes. In contrast, the durations of the bursts are always the same.
The researchers found that the characteristic bursting behavior can only be explained by assuming LexA undergoes rapid and transient depletions in concentration every time DNA damage occurs. These dips in concentration are so fast that, for some target genes, they may pass without the transcription factor ever unbinding from the DNA. “If LexA binds very strongly to a gene’s regulatory region, its concentration must remain low for a longer period before the gene is activated,” says van Nimwegen. “Each gene responds to DNA damage in its own unique way. Some genes only respond to severe, while others respond even to minor damage.” Depending on how strongly LexA binds, genes respond more or less sensitively to the same signal.
Gene regulation more complex than expected
These findings support earlier work by the research group showing that bacteria exploit molecular noise to respond more flexibly and efficiently to their environment. What may appear to be randomness could in fact be an evolutionary advantage, enabling bacteria to survive sudden environmental changes or threats such as antibiotics.
“Our work challenges the conventional understanding of how genes are regulated in bacteria,” says van Nimwegen. “Fast transcription factor fluctuations may well be ubiquitous in bacterial gene regulation, causing diverse responses across targets of the same regulator. This diversity in responses has likely been tuned by evolution, representing a sophisticated regulatory strategy.”
Original publication:
Luca Galbusera, Gwendoline Bellement-Theroue, Thomas Julou, and Erik van Nimwegen. Rapid transcription factor fluctuations drive nonequilibrium gene regulatory dynamics in bacteria. PRX Life; published online 18 July 2025
Contact: Communications; Katrin Bühler