Main Content
Quantitative biology of microbes
We combine quantitative experiments and theoretical models to elucidate how robust properties seen in bacterial populations emerge from seemingly stochastic dynamics observed in individual cells.
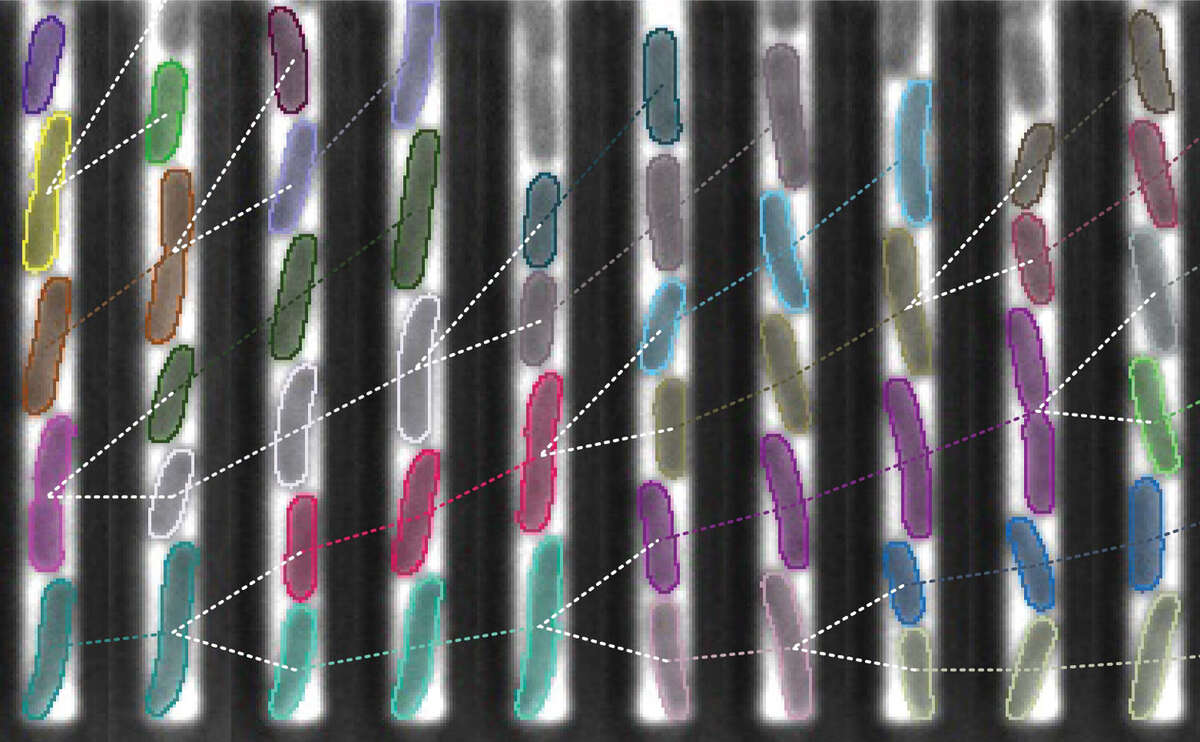
Because bacteria are small and chemical reactions involved in gene expression are inherently stochastic, different individual bacteria display different properties – so called phenotypes – even when they are genetically identical and exposed to the same environment. We study the mechanistic origins as well as the functional and evolutionary implications of this phenotypic variability. To this end, our group develops new tools to study bacteria with single-cell resolution, in particular using microfluidics and image analysis.
Learning from fluctuations
Although intracellular processes are noisy, many events of the cell cycle must be tightly controlled in order to sustain growth and division. Analyzing the fluctuations between cells in constant environments allows us to learn more about how different aspects of bacterial physiology – for instance cell division and DNA replication – are coordinated. In parallel, we study how robust gene regulation can be achieved in fluctuating environments, despite noise in gene expression.
Coping with stress
Our group also studies responses to stress where rare phenotypes are expected to play a major role. First, when bacteria are starving, we aim at understanding how cellular physiology is reorganized, and in particular what drives the variability of the time required for bacteria to restart growing. But also, when bacteria are treated with antibiotics, we want to characterize the non-genetic determinants of sensitivity to drugs.