The forefather of phylogenetic trees can be found in Charles Darwin's revolutionary work "On the Origin of Species". These trees provide information about the evolution and relationships of species, from simple cells to complex organisms. The length of the branches that separate two organisms in a phylogenetic tree reflects how much time has passed since these two organisms had a common ancestor. In the case of bacteria, such phylogenetic trees are nowadays mainly reconstructed from differences in the genome. The more similar their genomes, the more recently they are assumed to have had a common ancestor.
The scientific consensus that phylogenetic trees of bacteria depict how they are related, however, is now being challenged by the team led by Prof. Erik van Nimwegen at the Biozentrum of the University of Basel. “We have been working on this project for almost 10 years. And now, we believe we have finally cracked a major puzzle about the evolution of bacterial genomes,” says van Nimwegen. “In a nutshell, we have discovered that phylogenetic trees do not represent what most people have been assuming they do.”
Phylogenetic trees do not show ancestry
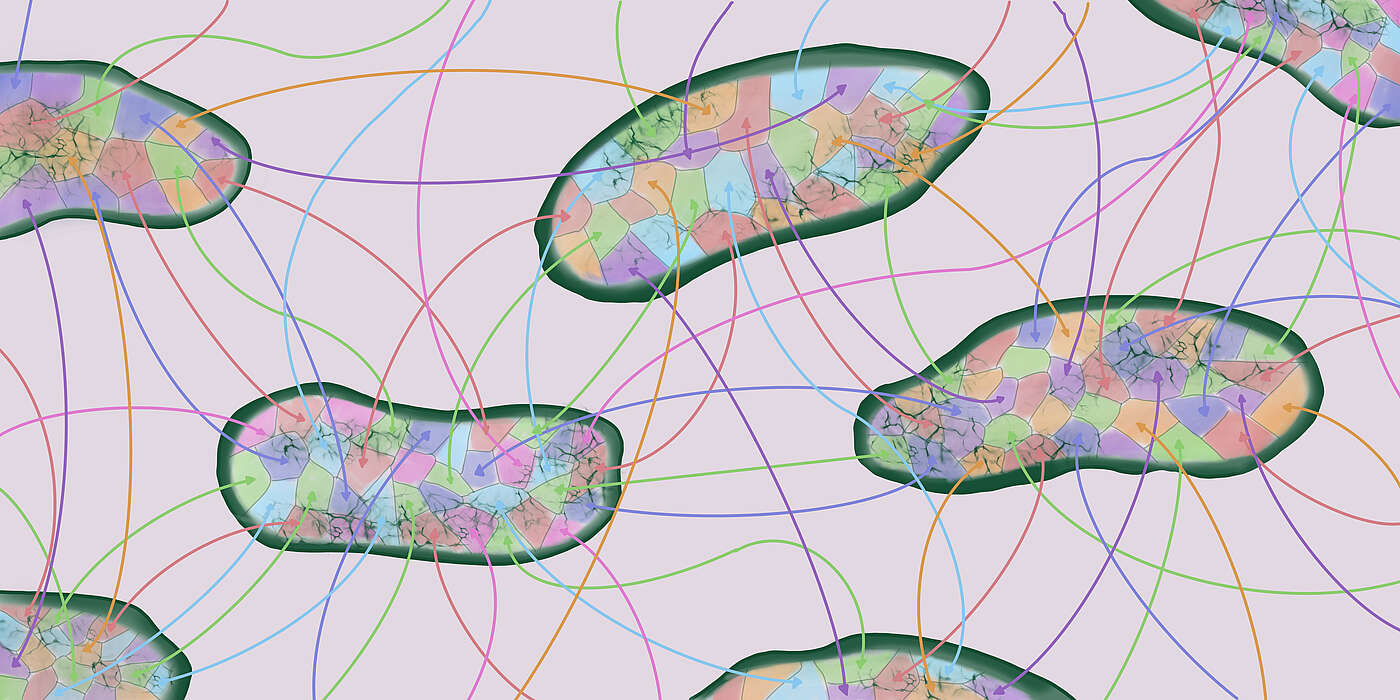
To reconstruct a phylogenetic tree from genome sequences, one has to assume that genetic mutations are passed from mother to daughter cell at division. Although bacteria do indeed pass their genes on from one generation to the next, this is not the only way. Bacteria can also transfer genes to each other. Via this so-called `horizontal’ gene exchange, for example, antibiotic resistance can be transferred to another pathogen.
Although the existence of horizontal gene transfer has long been appreciated, so far researchers have assumed that this process does not significantly affect tree reconstruction. However, using new analysis methods, the new research show this is mistaken and based on the incorrect presupposition that all bacteria within a species exchange DNA at approximately equal rates. However, it turns out that different bacteria exchange DNA at very different rates.
Bacterial genome evolution based on gene exchange
In their study recently published in “eLife”, the researchers find that phylogenetic trees mainly reflect the relative rates with which different strains have been exchanging genes, rather than tracing their evolution. “Bacteria exchange so much DNA with each other that the evolution of their genome is completely dominated by this gene transfer,” explains van Nimwegen. “Thus, the phylogenic trees built from genome sequences do not reflect when bacteria last had a common ancestor but rather how frequently they have exchanged genetic material with each other.”
Bacterial genomes do not evolve along a tree
Instead of genomic changes mainly occurring along the diverging branches of a tree, changes in bacterial genomes are predominantly introduced along many lateral connections between the branches, representing the transfer of single or multiple genes between the bacteria. “Our results change the way we need to think about bacterial genome evolution,” says van Nimwegen. “They imply that the genome data and the phylogenies reconstructed based on this data have been misinterpreted for more than twenty years.” Finally, the work also raises a challenge to evolutionary theorists, since it shows new models are needed to capture how bacterial genomes evolve.
Original publication:
Thomas Sakoparnig, Chris Field, Erik van Nimwegen. Whole genome phylogenies reflect the distributions of recombination rates for many bacterial species. eLife (2021)
Contact: Communications, Katrin Bühler